Computational Genomics Research Program
Head of the Program: Julio Collado-Vides, Ph.D.
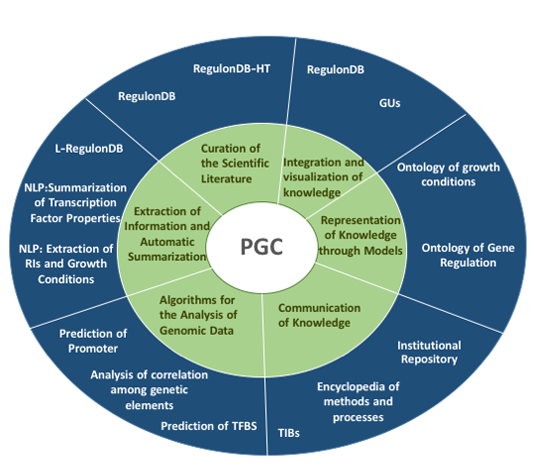
Research Topics and projects
Curation of the Scientific Literature
The typical curation process consists on reading scientific articles to extract information, in our case information related to E. coli K-12 transcriptional regulation, and organizing it in a database such as RegulonDB. One of the most important aims in the lab is to keep RegulonDB’s curation as up-to-date as possible.
Currently, we are developing a platform to curate digitally, which will avoid typographic mistakes and will lessen the amount of paper used for printing articles. The curator will mark a word in an article and, by selecting a capture form textbox, the word will automatically be copied.
We have also developed assisted curation, which makes use of text-mining techniques to filter information and detect phrases that contain the information we are interested in curating. So far, we have developed filters for annotating Regulatory Interactions (RIs) and growth conditions that have an impact on RIs.
Ongoing projects
• Common Curation
• Assisted Curation
• Curation of RegulonDB
• Curation of Ecocyc
• Curation of RegulonDB-HT
Algorithms for the Analysis of Genomic Data
RegulonDB contains big amounts of reliable information on genetic regulation of Escherichia coli K-12. We are interested in integrating data to discover new knowledge.
So far, we have developed algorithms for the search and discovery of Transcription Factor (TF) binding sites, promoter prediction and identification of co-expressed genes from expression profiles. Results from these efforts have enriched RegulonDB.
Ongoing projects
• Development of algorithms for prediction of TF binding-sites.
• Development of algorithms to predict orthologs in bacteria.
• Development of algorithms for predicting Transcription Start Sites (TSS’s).
• Development of algorithms to analyze gene co-regulation.
Integration and visualization of knowledge
We are aware that the nature of knowledge is representation and believe that what RegulonDB has done through the years is to accelerate the access to data and information on the regulation of transcription initiation in E. coli K-12. The challenge that guide us is:
How to implement representations that could accelerate access to knowledge?
This is not only a philosophical question. In practice we consider "knowledge" as every sentence that is contained in the corpus of papers supporting the data and information in RegulonDB that conveys data not yet encoded in RegulonDB. We estimate that 70 to 75% of sentences have not been encoded yet.
Ongoing Projects
• RegulonDB database
• Genetic Sensory Response Units
• L-RegulonDB
• Multigenomic database on gene regulation
• RegulonDB-HT
Representation of Knowledge through Models
RegulonDB hosts a myriad of information on transcriptional regulation. There are several projects in the laboratory that seek to improve communication of information so that the community can easily obtain a global perspective of available data, generate new hypothesis or elucidate unknown design principles.
Projects in this aim have two main focuses:
1) The categorization of data and its relationships through the design of ontologies and relational models.
2) The development of new concepts that aid in integrating data from a different perspective in order to obtain new knowledge.
Ongoing Projects
• Genetic Sensory Response Units
• Mathematical modeling of Transcriptional Regulation
• Ontology of RegulonDB entities
• Ontology of Growth Conditions
Communication of Knowledge
The Computational Genomics Research Program of the Center for Genomic Sciences pioneered Bioinformatics in Mexico; for years it has compiled, integrated and represented knowledge on genetic regulation. Nowadays, we have taken one of the main objectives of UNAM as our own: the compromise to communicate and spread knowledge.
Our interest is to facilitate the understanding of genomics and bioinformatics by society in general. We aim to provide ordered and standardized access to the information we generate through an institutional repository, and to provide training for the community interested in genomics and bioinformatics.
Ongoing Projects
• Institutional Repository of Genomic Knowledge
• Encyclopedia of Methods and Processes
• International Workshops on Bioinformatics
Extraction of Information from Scientific Articles and Automatic Summarization
Information Extraction. Typically, information extraction (IE) gathers a set of structured data that describe an event from an unstructured data source (documents, videos or images). In the biomedical field IE has been used to extract protein-protein and gene-gene interactions from collections of scientific articles. In the lab, we are particularly interested in using IE to extract regulatory interactions between transcription factors and transcription units, along with the growth condition in which they happen.
Currently, we use two IE methods to extract regulatory interactions: one based on syntactic rules and the other on filtering lists of words. Extracting interactions is complicated given that there are many different ways to phrase them; we are making efforts to enhance existing methods and efficiently retrieve more interactions and conditions. New methods will allow us to extract information from collections of articles concerning different microorganisms. The ultimate goal of this line of research is to assemble a complete transcriptional regulatory network from literature alone.
Automatic Summarization. Automatic summarization seeks to generate a shorter version of a document, or a collection of documents, that maintains relevant information. These methods can be used to obtain a summary of the principal characteristics of a biological entity, a disease or a treatment. In the lab, we are aiming to automatically obtain summaries of features of transcription factors from the scientific literature.
Ongoing projects
• Information Extraction
• Automatic summarization of TFs
• L-RegulonDB
• Multigenomic Database
Algorithms for the prediction of Transcription Factor Binding Sites (TFBS).
Introduction
Genetic regulatory systems have always been of interest to molecular biologists however, before the genomic era the characterization of one regulatory site was a time-consuming process. Nowadays, genomic techniques provide numerous binding sites in just one experiment, but the number of false positives can be very high and bioinformatic approaches are needed for the analysis and interpretation of results. Furthermore, through bioinformatics it is possible to use experimental information to create models that predict new regulatory sites.
Main goal
Implement algorithms and methodologies for the search and discovery of patterns in binding sites of transcriptional regulators in order to reduce false positives.
Description
One of the major caveats in pattern-matching approaches used to find new binding sites is the high number of false positives. In order to circumvent this problem, we integrate two methods to our processes:
• Evaluation of the quality of computational models known as Positions Specific Scoring Matrices (PSSMs), used to predict transcription factor binding sites. As a result of this study we propose the utilization of a multigenomic approach to improve PSSMs and make them available as part of RegulonDB.
• Utilization of multigenomic information, derived from the conservations of regulatory interactions in closely related genomes, as a way to reduce the quantity of false positives.
There have been other methods and approaches aimed at improving the quality of similar predictions, however, most of them require a lot of experimental information that is only available for few organisms. In the laboratory, we aim not only to discover new E. coli K12 binding sites but also to develop methods that can be applied to any organism with an annotated genome.
Project leader and participants
Alejandra Medina Rivera
Jacques van Helden
References
[1] Medina-Rivera A, Abreu-Goodger C, Thomas-Chollier M, Salgado H, Collado-Vides J, van Helden J. Theoretical and empirical quality assessment of transcription factor-binding motifs. Nucleic Acids Res. 2011 Feb;39(3):808-24. doi:10.1093/nar/gkq710. PubMed PMID: 20923783; PubMed Central PMCID: PMC3035439.
[2] Medina-Rivera A, Defrance M, Sand O, Herrmann C, Castro-Mondragon JA, Delerce J, Jaeger S, Blanchet C, Vincens P, Caron C, Staines DM, Contreras-Moreira B, Artufel M, Charbonnier-Khamvongsa L, Hernandez C, Thieffry D, Thomas-Chollier M, van Helden J. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015 Jul 1;43(W1):W50-6. doi: 10.1093/nar/gkv362. PubMed PMID: 25904632; PubMed Central PMCID: PMC4489296.
Algorithms for the analysis of correlation among genetic elements.
Introduction
Despite the great strides made by the RegulonDB team to integrate data from the literature into the most complete Transcriptional Regulatory Network of an organism, 2919 genes of a total of 4650 are not present in the E. coli network. We have developed a new line of research in the laboratory that uses computational approaches, genomic data and novel mechanisms of transcriptional regulation as an alternative for identifying missing interactions. This way, RegulonDB not only aims at exhaustive literature curation but also at generating new knowledge from publicly available genomic datasets.
Main goal
Develop computational algorithms for the global analysis of gene expression datasets (various experiments in several conditions) to identify new transcriptional regulatory interactions in E. coli, and assemble pipelines that increase the reproducibility of our results.
Description
The project implements computational algorithms based on machine-learning that analyse gene expression data to obtain new knowledge of transcriptional regulation [Martínez et al 2016].
The main sources of information are:
a) RegulonDB. Database that contains experimental information about transcriptional regulation such as regulons, transcription units and operons; the basic concepts that represent groups of genes that simultaneously respond to a stimulus.
b) M3D and COLOMBOS. Databases that contain gene expression datasets of E. coli and other organisms across several physiological conditions.
Two computational algorithms based on clustering have been used; they consider gene expression values across multiple conditions and group genes that have a similar expression profile. The resulting groups have been validated by control sets that include regulons, transcription units and operons, as annotated in RegulonDB.
These techniques have allowed us to infer regulatory interactions of genes without functional annotation through their membership to highly correlated groups.
Project leader
Mishael Sánchez-Pérez
Participants:
Socorro Gama-Castro
Alberto Santos-Zavaleta
Cecilia Ishida
David Velázquez
Irma Martínez-Flores
Victor Bustamante
Thesis and collaboration openings:
Undergraduate
Postgraduate
Internships
Data Analysis collaboration
References
[1] Martinez et al 2016. In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Irma Martínez-Flores, Deyanira Pérez-Morales, Mishael Sánchez-Pérez, Claudia C. Paredes, Julio Collado-Vides, Heladia Salgado & Víctor H. Bustamante. Scientific Reports 6, Article number: 37858 (2016) doi:10.1038/srep37858.
Curation of scientific articles in RegulonDB and EcoCyc
Introduction
RegulonDB is a database containing information on transcriptional regulation of E. coli K-12; it arose in 1991 with a collection of transcriptional regulator DNA binding sites, which was published in the article "Control site location and transcriptional regulation in Escherichia coli" (Collado-Vides et al., 1991). The database was first published in 1998 through the article "RegulonDB: a database on transcriptional regulation in Escherichia coli" (Huerta AM et al., 1998). RegulonDB is a high-quality database whose information is manually curated and where each object’s record contains the evidence that supports its existence and the source or reference from which the information for each object was obtained.
Main goals
1. To keep the curation of transcriptional regulation up to date.
2. To develop a digital system for daily curation.
3. To maintain assisted curation of information regarding growth conditions affecting genetic expression.
Description
The traditional curation process consists of the reading of scientific articles to extract information, in our case, on the regulation that affects genetic expression in E. coli K-12, and to annotate this information in the databases RegulonDB and EcoCyc. The objective of our group is to release new versions of the two databases four times a year and keep them as current as possible.
The curation process begins with a search for articles in PubMed, for which we use a set of keywords related to the topic of transcriptional regulation. The result of the search is a set of abstracts of articles, which are read in order to select all those that contain relevant information. Subsequently, complete articles are obtained in order to read them. The team of biocurators follows a unified set of curation criteria or guidelines, which has expanded as our experience has increased. The data extracted from the complete articles are added to EcoCyc through capture forms. These data are then transferred from EcoCyc to RegulonDB through an automated process, which is executed prior to each release. It should be mentioned that RegulonDB contains some data that are not shared with EcoCyc, and such data are annotated independently for later addition to RegulonDB.
Quality control of aggregate data is automatically performed through consistency checks. The questions, observations, and comments related to genetic regulation that are sent by the users to both databases are answered by our team.
Although there is a careful manual curation process, it is not free from human error. To avoid such errors, we have started the project for the development of a digital curation platform, which will integrate different computational tools to carry out the curation. With the use of this new platform, we will also reduce the amount of paper used in printing. The platform will allow the curator to interact with the digital version of the article and the forms of capture, so that through the marking of texts in the article, such information can automatically be retained in the forms of capture. It is envisaged that development of this system will be completed by the end of 2017.
Another type of curation that we have developed is assisted curation, which uses text-mining techniques that allow the use of carefully designed filters to detect phrases in an article that have the information that interests us. This process is useful and efficient in allowing curation directed towards a specific element of RegulonDB. So far, we have developed filters for annotation of Regulatory Interactions (IRs) and for growth conditions that affect IRs or directly affect genes.
Project leader and participants
Project leaders:
Socorro Gama
Alberto Santos
Participants:
David Velázquez
Cecilia Ishida
Hilda Solano Lira
Jair Díaz Rodriguez
Fabio Rinaldi
Sara Martínez Luna
References
[1] Collado-Vides J, Magasanik B, Gralla JD.Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371-94. Review.
[2] Huerta AM, Salgado H, Thieffry D, Collado-Vides J. RegulonDB: a database on transcriptional regulation in Escherichia coli. Nucleic. Acids Res. 1998 Jan 1 (C;26(1):55-9.
[3] Gama-Castro, S., Salgado, H., Santos-Zavaleta, A., Ledezma-Tejeida, D., Muñiz-Rascado, L., García-Sotelo, J. S., … Collado-Vides, J. (2016). RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond . Nucleic Acids Research, 44(Database issue), D133–D143. http://doi.org/10.1093/nar/gkv1156
Curation of High-Throughput Scientific Literature
Introduction
The availability of articles from high throughput technology (HT) experiments focused on studying genetic regulation has been augmented through the past of the years. This type of experiments could not been threated as classical experiment for gathering biological knowledge from them. So the need of understand and extract this types of data is a challenge now a days. The biological need from gathering information from classical and novel experiments of the regulatory network from Escherichia coli is fundamental requirement, that will guide us to understand and generate knowledge of this gold standard network.
Objectives
Generate a conjunct of methods that could lead us to incorporate the datasets obtained of HT experiments on a novel database (RegulonDBHT.
• Creation of a pipeline to process raw data and extract relevant information from them in a pre-established format.
• Curation and extraction of processed data found in supplementary material or tables of scientific articles of HT.
• Generation of different pipelines that allow the biological inference of regulatory relationships (e.g. direct regulation, indirect regulation).
Description
The HT experiments include different type of technologies like ChIP (ChIP-ChIP, ChIP-seq, ChiP-exo), genomic SELEX, or transcriptome (Microarrays, RNA-seq). We have start curating datasets from genomic Selex (CRP, LeuO, H-NS) and ChIP-exo (GadE, GadW, GadX, OxyR, SoxS and SoxR), which have been uploaded to RegulonDB database on the next URL: http://regulondb.ccg.unam.mx/menu/download/high_throughput_datasets/index.jsp
We plan to extract all the articles from HT literature on Escherichia coli K-12 extracting raw data and processed data. From 60 articles related to High throughput (HT) on E-coli K-12, approximately 20 have raw data deposited in GEO and / or Array Express, with different technologies such as: ChIP-chip, ChIP-seq, ChIP-exo, RNA-seq, microarray and 5'RACE. These articles will generate data for TFs such as OxyR, SoxR, SoxS, Fur, RyhB, NsrR, FNR, H-NS, IHF, ArgR, GadE, GadW, GadX, Fis, PurR, Lrp, TrpR; In addition to the sigma factor RpoD (sigma70) and the Rho termination factor. We hope to be able to also include transcription start data (TSS).
Raw Data Curation and Extraction
The Raw data will be extracted from GEO and Array express databases. This job will be done with semiautomatic scripts and in an automatically option that is incorporated in our pipeline. We plan to re-analyze this dataset in a standardize and homologate way to ensure all of them are processed in the same conditions to for data reproducibility. The plan is that the result from the raw data analysis contains a general format to upload them as tracks or dataset to RegulonDB. We will extract all the related information of each dataset to document in a meta-curation information table that contains all the experimental condition and the related information of the experiment.
Processed data Curation and Extraction
This type of dataset will be gathered from the main articles or their supplementary material. The data will be extracted and saved in a standard format for the processing and incorporation in other pipelines. All extracted data will be treated as a processed result joining to the evidence the corresponding meta-information about the experiment or conditions of the corresponding dataset
Data Visualization and Storage
For the visualization of the data in tracks, it is suggested that they be displayed in a circular way, the peaks are shown and they can be hyperlinked to the information related information. Multi tracks will have different categories such as shared conditions or TF that react to the same condition.
There exist some fields that all the raw data must contains: TF / gene / peak length / Peak central position / Methodology / Algorithm / Evidence (Verena) / Reference.
When RNA-seq or microarrays experiments are included in the same article, the function or effect (indirect) of TF on genes with altered expression will be associated.
Project leader and participants
Responsables:
Alberto Santos
Mishael Sánchez
Participantes:
David Velázquez
Mishael Sánchez
Alberto Santos
Pedro A. Hernandez
References
[1] Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keleş S, Landick R, Kiley PJ. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 2013 Jun;9(6): e1003565. doi: 10.1371/journal.pgen.1003565. PubMed PMID: 23818864; PubMed Central PMCID: PMC3688515.
[2] Myers et al. 2013 Myers, K. S., Yan, H., Ong, I. M., et al. (2013) PLoS Genet., 9, 11–13, Genome-scale Analysis of Escherichia coli FNR Reveals Complex Features of Transcription Factor Binding.
Encyclopedia of methods and processes
Introduction
Nowadays, we are experiencing a revolution in the man-information interaction due to the fast-paced progress on Information and Communications Technologies (ICTs). Access to information is taken for granted. However, it has also become harder for the common user to recognize validated information.
This project is located at the beginning of the processing of knowledge by committing to accelerate the availability, not of information, as computers need it, but of knowledge as it is required by a human mind when exercising its capacity to understand.
The challenge of the project is to offer an alternative that maximizes knowledge understanding. It is important to provide reliable knowledge that has gone through an editing process that guarantees its reliability. The hypothesis is that all knowledge can be expressed in an easy way to understand for non-expert people and gradually going forward to more specialized and technical levels.
Main goals
This project of an Encyclopedia of Methods and Processes is a module aligned with the mission we have around the verb "to understand". Nowadays, the project includes texts in spanish, our long-term goal is to facilitate understanding for everyone.
The aim is to build an accessible online Encyclopedia of Methods and their integration in Processes in spanish, which together with strategies of knowledge management, will allow the reader to understand the basics of human genomics. In this project the scope is limited to scientific methods of the genomic sciences and bioinformatics, emphasizing those involved in personalized genomics.
Specific goals
1. To build of the first prototype of the encyclopedia.
2. To perform a market survey and a business proposal. The encyclopedia will be an additional module in the Conogasi system, to be offered in a future version.
3. To define the commercial strategy and legal basis of the company and services..
4. To define and document the processes that confer the robustness of the business project, so that we could launch the project at national level.
Project leader and participants
Leader: Julio Collado Vides
Participants:
• Marcela Castillo Figa
• Heladia Salgado Osorio
• Alejandra Cristina López Fuentes
• Gerardo Salgado Osorio
Other contributors
• Shirley Alquicira
• Kevin Alquicira
• Liliana Porrón
• José Alquicira
• Berenice Jiménez
• Alberto Checa
• Cecilia Ishida
• Luis Olarte
• Jesús Alvarado
• Omar Alva
• Masami Ando
Institutional Repository of Genomic Knowledge
Introduction
Genomic Science is undergoing a revolution derived from the technological advances that have facilitated the acquisition of massive amounts of data. The current challenge is to access, process, and integrate the information and knowledge generated.
In this project we aim to make information nationally and internationally accessible, public, structured and adequately commented while abiding by the interoperability rules required for national repositories of microbial genomics.
Main goals
The main goal is to create an institutional repository of interpretable elements of complete genomes. We will start a pilot project, spanning 6 months divided in two stages, with the orthology relationships among all the available microbial genes. We will collaborate with Dr. Gabriel Moreno-Hagelsieb, who has spent years identifying the relationships. (Ward N. and Moreno-Hagelsieb G. 2014; Vey G. and Moreno-Hagelsieb G. 2010; Moreno-Hagelsieb G. and Latimer K. 2008). The pilot project will be later enriched with other elements of complete microbial genomes, and eventually of eukaryotic genomes, especially human. This is a long-term project that will have national and international impact.
The specific goals of the pilot project are:
1. To obtain and configure the computational infrastructure needed for the repository.
2. To define standards of documents and processes to homogenize the data that will be deposited.
3. To design the structure of the repository.
4. To make available the orthology relationships of the genes in the 2785 microbial genomes available to date.
Project leader and participants
Gabriel Moreno Halgelsieb
Heladia Salgado Osorio
Gerardo Salgado Osorio
Shirley Alquicira Hernández
Support
Víctor del Moral Chávez
Romualdo Zayas Lagunas
Description of information fluxes in E. coli
Introduction
The elucidation of the Transcriptional Regulatory Network of Escherichia coli K12 made possible the analysis of genetic circuits that dictate the information 1–5 processing capabilities of the cell, however, little is still known about the physiological effect of those circuits. Transcription factors are central to information processing given their ability to allosterically bind to an effector, or the end-product of a signalling pathway, that reflects changes in the environment. The formation of the TF-effector complex alters the TF DNA-binding ability, which in turn activates or represses a defined group of genes termed “regulon”. Physiologically, this set of genes will give rise to proteins that together will orchestrate a metabolic response appropriate to the presence of the initial signalling molecule.
Signal-response processes take place countless times during a cell’s lifespan and the mechanisms underlying them provide the ability to recognize and adapt to changes in the environment.
Main goals
Describe the information processing units encoded in Escherichia coli K-12 and identify their general properties.
a) Use a data-driven approach to assemble a GENSOR Unit for each of the 189 local transcription factors present in RegulonDB.
b) Identify presence of feedback in each GENSOR Unit.
c) Quantify the functional homogeneity of each GENSOR Unit and identify global patterns.
Description
A GENSOR Unit is composed of 4 elements:
a) Signal; the metabolite that communicates a change in the internal or external environment.
b) Processing of the signal; in the case of transcription factors this component refers to the conversion of the signal into the effector that is recognized by the transcription factor.
c) Genetic switch; activation or repression of a group of genes.
d) Response; action derived from the coordinated action of the gene products whose expression has been altered by the transcription factor.
Currently, 189 GENSOR Units have been assembled through a semi-automatic approach that retrieves the necessary elements from 2 databases that include experimentally validated data. The next step is to analyse them in order to identify general principles used by the cell to detect and process information. GENSOR Units have a twofold goal in the long term:
(1) Provide a tool for analysis, not only of individual genes, but for their interactions, the signals that prompt them and the response they generate.
(2) Provide a framework for the integration of different levels of cellular organization to eventually obtain a cellular map where it would be possible to predict cellular behaviours in silico from the presence of combinations of signals.
Project leader and Participants
PhD project of: Daniela E. Ledezma-Tejeida
Cecilia Ishida
References
[1] Thomas, R. & D’Ari, R. Biological feedback. (CRC Press, 1990).
[2] Kauffman, S. A. The origins of order : self-organization and selection in evolution. (Oxford University Press, 1993).
[3] Gerosa, L. & Sauer, U. Regulation and control of metabolic fluxes in microbes. Curr. Opin. Biotechnol. 22, 566–75 (2011).
[4] Savageau, M. A. Design principles for elementary gene circuits: Elements, methods, and examples. Chaos 11, 142–159 (2001).
[5] Savageau, M. A. Biochemical Systems Analysis. A Study of Function and Design in Molecular Biology. (Addison-Wesley Publishing Company, 1976).
[6] Daniela Ledezma-Tejeida, Cecilia Ishida, Julio Collado-Vides. Genome-wide mapping of transcriptional regulation and metabolism describes information-processing units in Escherichia coli (submitted)
High-Throughput Literature Curation of Genetic Regulation in Bacterial Models
Introduction
In order to cope with the massive increase on information availability in all fields of knowledge, particularly in genomics, we are proposing new reading strategies based on Natural Language Processing techniques. The main goal is to use Semantic Similarity to provide the reader with a different approach for recearching a specific topic, allowing him to locate statements in other publications that convey the same or very similar ideas. This tool could be particularly helpful in the curation of scientific literature.
Main Goals
1. Develop a new system for navigation of scientific articles at the level of sentences.
2. Link sentences from scientific articles to the information stored in the RegulonDB database so that one can access the figures and tables in RegulonDB from articles on gene regulation.
3. Integrate in the system a navigable ontology about genetic regulation to classify scientific articles and their individual paragraphs.
Description
Nowadays we are facing a big challenge in terms of the amount of data and information that is being produced in several knowledge fields, and biological science is no exception. In genomics, the amount of data and information contained in publications is becoming so huge that to be up to date on any given topic we need assistance.
For this reason, we are developing techniques to read and follow ideas beyond the context of the publication where we first found them. The main goal is to provide the reader with a different approach for researching a specific topic, allowing him to locate statements in other publications that convey the same or very similar ideas. This could be a stepping stone towards an augmented reality in the context of Natural Language.
Our approach is based on the use of NLP techniques to measure semantic similarity among phrases and then select the strongest semantic links. The semantic similarity module is on continuous improvement, currently it combines techniques based on distributional semantics and some syntactic features, the latter will be expanded in later versions.
This reading approach can be applied to any field where people need to read publications in order to conduct research, but can be particularly helpful in scientific curation. In the curation process, the experts read one by one a set of topic-related articles to annotate relevant information. This technique works well in the sense that relevant information is identified but having to read the whole articles sequentially is very time-consuming. By using semantic links the experts could jump from publication to publication finding support or counter evidence in a simpler, more efficient way.
Project Leader and Participants
Project leader: Oscar Lithgow
Participants: Oscar Lithgow, Ale López
Automatic Extraction of Regulatory Interactions and Growth Conditions
Introduction
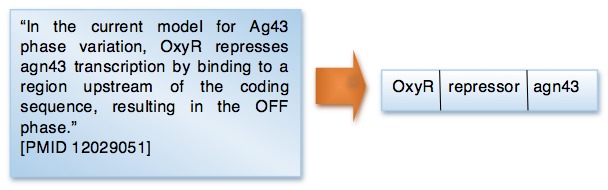
One of the products generated by the RegulonDB curation process of scientific articles is the transcriptional regulatory network (http://regulondb.ccg.unam.mx/menu/download/datasets/index.jsp). This network is formed by regulatory interactions between transcription factors (TFs) and genes or transcription units (TUs). In this project we are working on methods to these interactions and their associated growth conditions. Currently, we are interested in three types of interactions: activation, repression, and regulation. The following figure shows the regulatory interaction (right) that we would expect to obtain from a sentence in a scientific article (left).
Main goal
To automatically extract regulatory interactions between TFs and genes or TUs from biomedical literature.
Specific goals
1. To compare two approaches previously developed at the lab for extracting regulatory networks [1,2] from the E. coli network through standard metrics of information retrieval.
2. To select one of the approaches or propose an alternative and apply it on new article collections.
3. To extract the transcriptional regulatory network of Salmonella typhimurium.
Description
The proposed method will receive a scientific article collection, as input, and will produce a set of regulatory interactions and their associated growth conditions.
Project leader and participants
Carlos-Francisco Méndez-Cruz (leader)
Socorro Gama-Castro
Cecilia Ishida-Gutiérrez
Ignacio Arroyo-Fernández
Karla Jazmín Sánchez Jerónimo
References
[1] Rodríguez-Penagos, C., Salgado, H., Martínez-Flores, I., and Collado-Vides, J. Automatic reconstruction of a bacterial regulatory network using Natural Language Processing. BMC Bioinformatics (2007), 8:293, doi:10.1186/1471-2105-8-293.
[2] Gama-Castro, S., Rinaldi, F., López-Fuentes, A. et al. Assisted curation of regulatory interactions and growth conditions of OxyR in E. coli K-12. Database (2014) Vol. 2014: article ID bau049; doi:10.1093/database/bau049.
Automatic Summarization of Transcription Factor Properties
Introduction
RegulonDB includes summaries of the properties of transcription factors (TFs), each integrates several pieces of knowledge obtained through a curation process.
The summaries are produced by extracting relevant information from several scientific articles that are referenced throughout the text. Currently, there are 177 summaries with 13 references in average and a median of 9. To access the summary of any given TF search the name of the TF in the RegulonDB homepage, for example CytR, and select the section [Regulon]; the display of a summary is shown in the next figure:
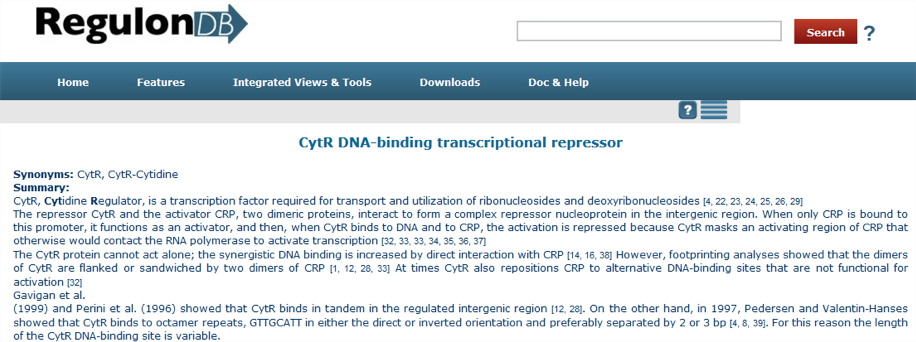
Summaries include the following information:
1. The meaning of the acronym and if the TF is a repressor, activator or dual transcriptional regulator.
2. The function in terms of its physiological role.
3. The growth conditions under which it is expressed.
4. The active or inactive conformation.
5. The number, name, and size of the structural domain constituting the TF.
6. Features of the binding site.
7. Information about its regulatory mechanism.
8. Information about its evolutionary features.
9. If the TF has other regulatory functions.
Goals
Automatic generation of summaries of the properties of transcription factors to contribute to the curation process of RegulonDB.
Description
Due to the accelerating growth of biomedical literature, to update these summaries and create new ones from different article collections became a demanding task for curators. Therefore, we propose to employ automatic summarization techniques to create these summaries.
Project leader and participants
PhD project of: Carlos-Francisco Méndez-Cruz
Socorro Gama-Castro
Citlalli Mejía-Almonte
Luis-José Muñiz-Rascado
Marco-Polo Castillo-Villalba
Alan Vladimir Godínez Plascencia
Cristian Jesús González Colín
Juan Antonio Blanchet Villezcas
Ontology of growth conditions affecting genetic expression in Escherichia coli.
Introduction
Cells have developed multiple mechanisms to detect signals from the environment and adjust their machinery to grow under different growth conditions. Most detection systems transduce and respond to a defined signal through the activation and/or repression of a particular set of genes that trigger or suppress a physiological response.
Since 2003, we have included in RegulonDB partial information on the growth conditions of the gene expression experiments reported in the literature. The manual curation of this information is a slow process, so we seek to implement semiautomatic curation to accelerate it.
Another database that also contains information on the growth conditions of gene expression experiments is COLOMBOS, which allows the exploration and analysis of complete summaries of bacterial gene expression experiments.
Main goals
1. Establish a controlled vocabulary for the annotation of growth conditions for the RegulonDB and COLOMBOS databases.
2. Create a specialized ontology for the growth conditions used in E. coli gene expression experiments.
Project leader and participants
Socorro Gama(leader)
Citlalli Mejía
Heladia Salgado
Kristof Engelen
Victor H. Tierrafría
Ontology of Gene Regulation
Introduction
RegulonDB is the primary database on E. coli K12 transcriptional regulation. One of the purposes of RegulonDB is to organize knowledge by means of formal computable models that enable faster access and processing. Relational databases allow the efficient storage and management of massive amounts of data. Although databases have a schema that models real world entities, they are not semantically relevant[1].
RegulonDB is closer to a representing knowledge than to mere data storage to the extent that we can represent true assertions about a domain of reality on which it is possible to apply logical reasoning to derive new knowledge; and can add semantics to the represented concepts, i.e. the context in which the concept makes sense.
Making possible the computational handling of all knowledge deposited in RegulonDB would strengthen the impact and relevance of the database. To this end, it is necessary to develop a computational model that can increase the number of formalized concepts about gene regulation, and precisely define their meanings to a level that provides a conceptual framework for the curation of new knowledge.
Aim
To make a formal description of the semantics of RegulonDB objects using ontology web language (owl).
Project leader and participants
PhD project of: Citlalli Mejía Almonte
References
[1] Martinez-Cruz, C., Blanco, I. J., & Vila, M. A. (2012). Ontologies versus relational databases: are they so different? A comparison. Artificial Intelligence Review, 38(4), 271-290.
[2] Blake, J. (2004). Bio-ontologies—fast and furious. Nature biotechnology, 22(6), 773-774.
[3] Blondé, W., Mironov, V., Venkatesan, A., Antezana, E., De Baets, B., & Kuiper, M. (2011). Reasoning with bio-ontologies: using relational closure rules to enable practical querying. Bioinformatics, 27(11), 1562-1568.
In search of innovation and quality (Processes).
Introduction
In recent years, information has become one of the most important strategic resources of any organization, and society in general. It is required to face the constant changes in social life and in the accelerated progress of information technologies (ITs). Unfortunately, it is common that the modern lifestyle does not allow time to store, organize and make the best use of information. Therefore, a current challenge is to identify what information we are producing and how to best manage it.
Research centers are no exception to the above-mentioned trend; project management initiatives under standardized methodologies or process models are almost nil, either due to lack of knowledge on tools to implement them or resistance to change. According to the CHAOS report, the most recognized report about success and failure of projects in the IT sector and produced by the Standish Group, there are critical factors that determine a successful project, of note are implementing a well-defined process model, and the use of methodologies that manage the model as well as the entire organization.
In the Computational Genomics Program we have made efforts for years to implement methodologies and models that aid in the management of projects and in the storage of the knowledge derived from them. In this project we work in a variety of methodologies and process models that allow us to produce better work products.
Main goal
To implement quality control and best-practice models to help the members of the laboratory to successfully execute their projects and store the knowledge and products generated in a repository or knowledge base.
Description
The Computational Genomics Program combines research projects from several fields. From its origins it has hosted RegulonDB, a database that has been the starting-point for graduate projects and new lines of research. The diversity of products generated at the laboratory is extraordinary, including information retrieved from scientific articles, specialized software development, development of algorithms for analysis of genomic data, definition of new concepts to explain gene regulation, theoretical modelling of the transcriptional regulatory network, etc.; all the above constantly producing in turn thesis, reports and scientific articles. All this information was not organized for many years, even though the necessity was already present.
In this project we focus on the implementation of models and development of tools necessary to build and maintain a knowledge base, and to evaluate its impact on the laboratory members.
Efforts to standardize project management took place since 2002, but it was until 2010 when the laboratory, through its software development unit, obtained a certification under the MoProsoft quality model. Nowadays we hold level 2 of the certification. This has increased the quality of the software generated and has positively impacted on the organization of all the activities that take place in the Computational Genomics Program.
The next step is to efficiently expand the culture of organization to all the projects in the laboratory.
Project leader and participants
Gerardo Salgado Osorio(leader)
Heladia Salgado Osorio
Shirley Alquicira Hernández
Liliana Porrón Sotelo
References
[1] CHAOS Report : https://www.standishgroup.com/store/services/pre-order-2016-chaos-report.html
International Workshops on Bioinformatics (IWB)
Introduction
In September 2001 Dr. Juan Ramón de la Fuente, former chancellor of UNAM, created the Bioinformatics National Node EMBNet (European Molecular Biology Network) – Mexico seeking to promote Bioinformatics in the country. Currently, it is based at the Center for Genomic Sciences (CCG-UNAM) and is promoted by the Computational Genomics Program.
EMBNet is a worldwide organization that brings together professionals who serve, support and maintain the growth of Bioinformatics in 30 countries. The Bioinformatics National Node (EMBNet-Mexico) pursues the same goals and maintains the philosophy of the global organization:
• To provide education and training in bioinformatics.
• To exploit network infrastructures.
• To investigate, develop and use public domain software.
• To assist research related to biotechnology and bioinformatics.
• To create bridges between commercial and academic sectors.
• To promote global cooperation through its community networks.
International Workshops on Bioinformatics (IWBs) are quality courses and workshops on frontier issues that address the aim of supporting and promoting Bioinformatics in Mexico.
Description
IWBs organization begins with the designation of an Executive Board to coordinate administrative and academic activities including diffusion of the workshops, evaluation and selection of infrastructure and logistics. Academics from the UNAM community are also invited to be part of the board.
The Center for Genomic Sciences (CCG-UNAM), the Undergraduate Program on Genomic Science (LCG-UNAM) and the Institute of Biotechnology (IBT-UNAM) provide the facilities, financial resources, computational infrastructure and food services.
From the first workshop in 2010 experts in the area, with high-skills and a love for teaching, have been sought to achieve high quality courses. It is important to note that all instructors, both national and international, have worked pro bono, reflecting their commitment to promote Bioinformatics in Mexico.
The results have been very satisfactory. The first edition in 2010 included a single course on "Introduction to Bioinformatics" for 40 students; the most recent event in January 2017 comprised 6 workshops (3 per week) with national and foreigners teachers, and was attended by more than 240 participants.
The IWBs are the most important activity of academic promotion carried out by the CCG through EMBNet-Mexico; they tend to the increasing demand on students and researchers of having bioinformatics skills.
Throughout the years, we have confirmed that great interest on bioinformatics courses and workshops exists in Mexico, and it is for this reason that we will keep organizing and promoting, alongside our collaborators. the IWBs
Project Leader and participants
Organizing Committee of the IWB 2017:
M.C. Romualdo Zayas Lagunas (CCG, UNAM)
Head of the Administrative subcommittee.
LCC Heladia Salgado Osorio (CCG, UNAM)
Head of the Academic Subcommittee.
Dr. Irma Martínez Flores (CCG, UNAM)
Head of the Evaluation Subcommittee
LCC Alfredo J. Hernández Álvarez (LCG, UNAM)
Head for the Subcommittee on Infrastructure and IT
Dr. Rosa María Gutiérrez Ríos (IBt, UNAM)
Head of the Logistics Subcommittee
Dr. Mishael Sánchez Pérez (CCG, UNAM)
Head of the Sub-Committee on Diffusion and Dissemination
MATI César A. Bonavides Martínez (CCG, UNAM)
Administrative Subcommittee
• Dr. Pablo Vinuesa Fleischmann (CCG, UNAM)
Academic Subcommittee
References
[1]. Website of the Bioinformatics National Node (EMBNet-Mexico)
http://www.nnb.unam.mx/
[2]. IWB website
http://congresos.nnb.unam.mx
| Responsible | Dr. Julio Collado Vides |
| Researcher(s) | Dr. Carlos Francisco Méndez Cruz |
| Academic Technician(s) |
LCG José Alquicira Hernánez, MTI Kevin Alquicira Hernández, MTI Shirley Alquicira Hernández, MTI César A. Bonavides Martínez, ISC Martín Jair Díaz Rodriguez, IE Víctor Manuel del Moral Chávez, MCB Socorro Gama Castro, LI Delfino García Alonso, DBC María Cecilia Ishida Gutiérrez, MTI Oscar William Lithgow Serrano, MSI Alejandra López Fuentes, Biol. Sara Berenice Martínez Luna, TMC Gabriel Martínez Posada, MTI Luis José Muñíz Rascado, ISC Luis Olarte Gervacio, ISC Vicente Osorio Mora, ISC Pablo Peña Loredo, II Liliana Porrón Sotelo, MAF Gerardo Salgado Osorio, LI Heladia Salgado Osorio, DCC Mishael Sánchez Pérez, MBT Alberto Santos Zavaleta, MTI Hilda Solano Lira Biol. Víctor Hugo Tierrafría Pulido, MCB David Alberto Velázquez Ramírez, MCC Romualdo Zayas Lagunas |
| Students |
PhD: ME Ignacio Arroyo Fernández MC Marco Polo Castillo Villalba LCG Daniela Elizabeth Ledezma Tejeida Bio. Citlalli Mejia Almonte Residents: LCG Juan Antonio Blanchet Villezcas ISC Gustavo San Luis Briseño LCG Alan Vladimir Godínez Plascencia LCG Cristian Jesús Gonzalez Colín ISC Francisco de Jesus Guadarrama Garcia |
| Administrative Assistant(s) | Concepción Hernández Lévaro |
| General Service Assistant(s) | David Castrejón Sánchez |
| top |









